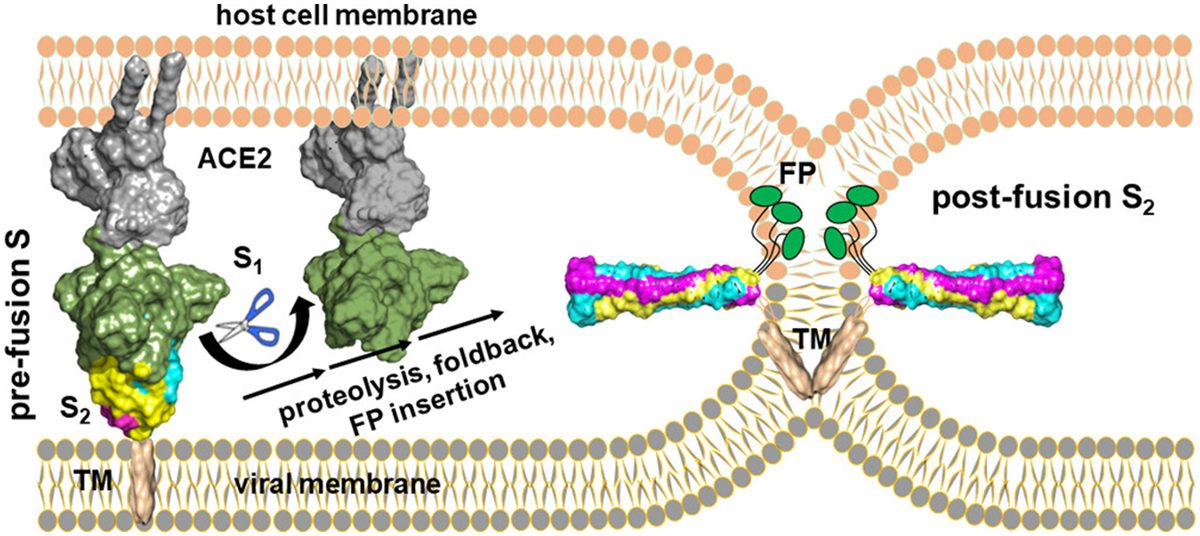
Virus Entry Steps
Project 1: Small Molecule Entry Inhibitors of Pandemic Viruses Abstract
Coronaviruses (CoVs), arenaviruses (Arv), flaviviruses (FLAVs), and filoviruses (FiVs) are enveloped viruses. An envelope can protect a virus's genetic material (i.e., the viral genome) during its life cycle when traveling between host cells.
During virus entry of these viruses, receptor binding and refolding of the glycoprotein protein embedded in the envelope, followed by lipid mixing, are three essential steps to release the viral genome. Therefore, inhibitors of any one of these three steps may be developed as effective antiviral drugs.
Our project has three aims. In our first aim, we will screen compounds that bind to the spike protein from SARS2 and the envelope protein of the Zika virus.
High-throughput screening targeting the protein refold formed by SARS2 spike protein (its glycoprotein on the envelope) will be carried out. In addition, virtual screens by AutoDock (a molecular modeling simulation software) or other computation methods will be conducted.
Our second aim will focus on discovering mechanisms of action of a successful. Finally, we will screen entry inhibitors for receptor binding inhibition with the SARS2 spike protein.
Entry inhibitors of other viruses will follow the same study approach.
In preliminary efforts, we have identified three inhibitors for SARS2. Mechanistic studies confirm that these inhibitors block membrane fusion during virus entry.
Ebola virus entry inhibitors are at an advanced stage and serve as a proof-of-concept for our strategy. We have identified a series of small molecule inhibitors targeting the Ebola virus with a novel mechanism. We will evaluate and adapt these for the broad-spectrum activity against other significant viruses and evaluate them in vivo efficacy in animal models.
We will develop a state-of-the-art approach to evaluate mutants that escape antiviral activities to aid inhibitor optimization.
In aim three, we will focus on in vivo efficacy. For SARS2, we will evaluate lead candidates for broad antiviral activities against multiple SARS2 and SARS isolates. Potent candidates will be evaluated in hamster and mouse models.