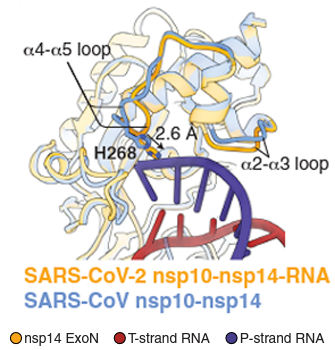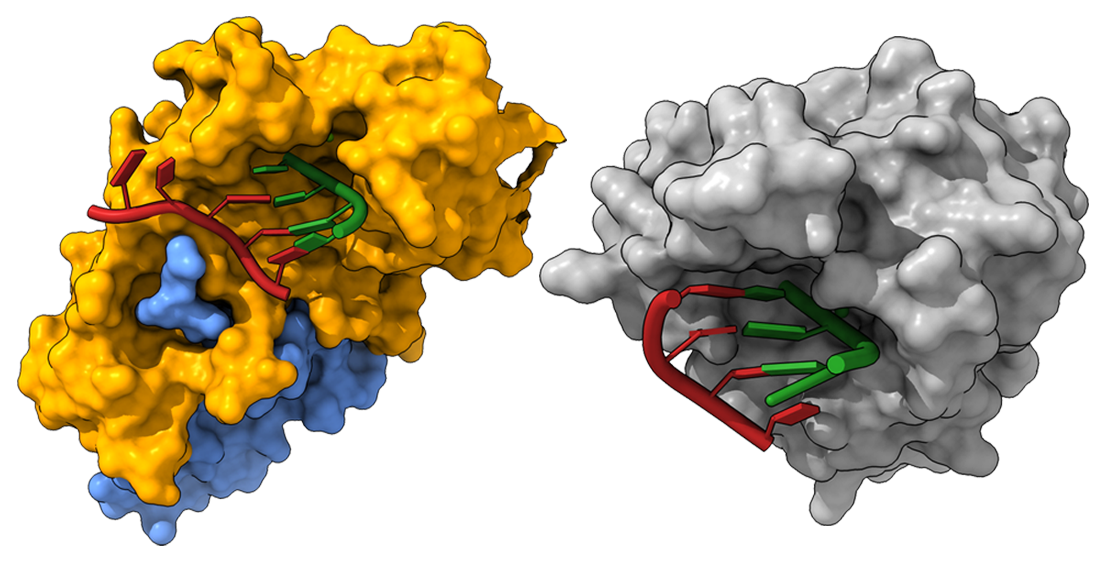
SARS-CoV-2 (left) and Lassa virus (right) ExoN engaging the RNA substrate
Image Credit: Yang Yang
Reference: (Left) Liu et al., Science (2021) Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. (Right) Jiang et al., JBC (2013) Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
Principal Investigator

Hideki Aihara, Ph.D
Deputy Investigator

Yuying Liang, Ph.D

Deputy Investigator

Hinh Ly, Ph.D

Yang Yang, Ph.D: Iowa State University

Daniel Harki, Ph.D: University of Minnesota, OSC

Robert Davey, Ph.D: Boston University, OSC

Martin Matzuk, M.D., Ph.D: Baylor University, OSC

Rommie Amaro, Ph.D: UC San Diego, OSC

Louis Scampavia, Ph.D: UF Scripps, OSC